What Makes This Invasive, Non-Native Reed Grass Thrive in the Wetlands?

Native and invasive Phragmites australis.Photo Credit: Dassanayake Lab
BATON ROUGE—The Mississippi River Delta is home to the world’s largest contiguous swath of Phragmites australis, or more commonly known as the common reed. But the plant that can grow to nearly 20 ft. and has been a critical component in stabilizing the state’s coastal erosion is not actually native to Louisiana—well, not entirely.
There are multiple P. australis genotypes. P. australis subspecies (ssp.) americanus is the native subspecies in the U.S. and Canada. However, Phragmites australis ssp. australis originated in central Europe and was subsequently introduced to the U.S. where it is now considered to be one of the most problematic invasive species in North America.
What has perplexed environmental researchers is the invasive ssp. australis has displayed capabilities beyond that of the native ssp. americanus in its ability to thrive in the wetlands, especially around the Great Lakes, often growing up to be much taller and denser, and in turn, disturbing the native ecosystem.
In a newly published study in Molecular Ecology and recently featured in an edition of The Scientist, LSU researchers collaborated with Tulane University and the U.S. Geological Survey to study the genomic bases of P. australis and to investigate what exactly makes the invasive reed grass subspecies thrive in wetlands, in comparison to its native counterpart. Samples were used from sites located around the Great Lakes region for this pioneering genomic study, though the plant can be found growing throughout North America.
“We are trying to understand the genomic basis for invasiveness in plants,” said Dong-Ha Oh, research assistant professor in the Dassanayake Lab in LSU’s Department of Biological Sciences and lead author of the paper.
This project resulted in the first genome reference for this globally recognized invasive plant that can be used by plant scientists studying evolution of invasive traits as well as scientists designing genetic-based strategies to manage invasive plants in conservation biology.
The study also included a comparison of gene expression data (comparative transcriptomics). When used with the newly assembled genome, it suggested that genes associated with pathogen and defense responses were highly expressed in the invasive subspecies continuously, while similar genes in the native subspecies were found at much lower expression levels and were only induced when there was a pathogen.
“We are seeing a built-in defense response in the invasive plants that is much higher than in the native plant,” said Maheshi Dassanayake, associate professor in the Department of Biological Sciences and a corresponding author of the paper. “For example, if we give both of these plants a pathogen and then test what happens, we see the native one acting drastically to respond to the attack, while the invasive one just doesn’t care because it always has its shields up.”
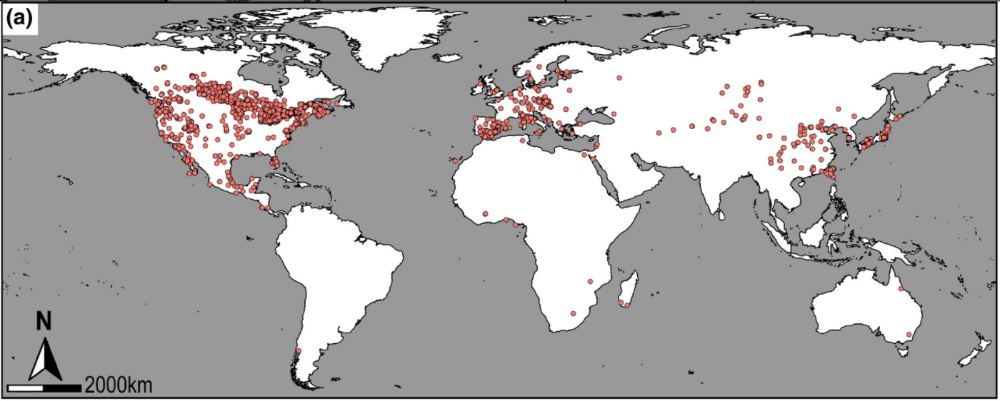
Reported global distribution of the invasive Phragmites australis ssp. australis.Photo Credit: Dassanayake Lab
Chathura Wijesinghege, a graduate student in the Dassanayake Lab contributed to this work by tracing the evolutionary history of Phragmites and closely related grasses. Dassanayake was invited to collaborate on an existing project between Tulane’s Keith Clay and USGS’s Kurt Kowalski that funded a genome project with a goal to design genetic control measures that can differentiate native subspecies from the invasive sub-species without causing unintentional damage to native fauna and flora.
“The USGS recognized the management need and initiated analysis of the genetic makeup of Phragmites as part of the new study,” said Kowalski. “This cutting-edge research provides a roadmap for further development of species-specific treatments to control invasive Phragmites and offers insights into how it compares to other grasses.”
The Dassanayake Lab analyzed the invasive plant’s genome using LSU’s High Performance Computing (HPC) services and revealed a unique history of genome-wide duplication events that likely provided novel genetic material for the divergence of the invasive and native subspecies. After identifying reference genes in the genome, the group looked at their expression in the native subspecies in comparison to the invasive.
“(This invasive reed subspecies) is destroying ecosystems that have been adapted to the native reeds, and (the USGS) wants to find out some biological solution that avoids the use of generic herbicides or labor intensive-mechanical removal,” said Oh.
“If we just leave it and perhaps in hundreds of years, the ecosystem may eventually adapt to this invasive species, but we may likely lose much of the local biodiversity in the meantime. So, plant biologists and conservation biologists can work together to find effective and sustainable solutions to control this problem before irreversible damage is observed to our native communities.”